NCERT Solutions for Class 12 Biology Chapter 11 Biotechnology: Principles and Processes
Biology for Class 12 solution of biology, we have included all the chapter questions. Biology Class 12 PDF Solutions will prove to be beneficial for the board exams as well as for the students who are preparing for NEET, UPCPMT and other Medical Entrance exam PDF.
biotechnology: principles and processes class 12 ncert solutions, biotechnology principles and processes class 12 notes, biotechnology principles and processes class 12 ncert solutions pdf download, biotechnology principles and processes class 12 questions and answers pdf, biotechnology principles and processes class 12 ncert pdf, download biotechnology principles and processes class 12 pdf, biotechnology principles and processes notes for neet, biotechnology and its applications class 12 ncert solutions
These Solutions are part of NCERT Biology 12 solution Here we have given NCERT Solutions for Class 12 Biology Chapter 11 Biotechnology : Principles and Processes
Chapter 11 Biotechnology: Principles and Processes
Question 1.
Make a chart (with diagrammatic representation) showing a restriction enzyme, the substrate DNA on which it acts, the site at which it cuts DNA and the product it produces.
Solution:
Name of the Restriction enzyme – Bam HI.
The substrate DNA on which it acts –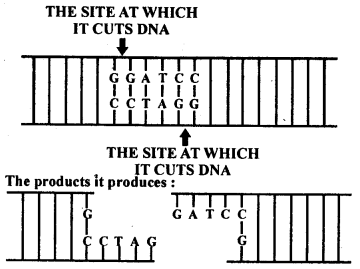
Question 2.
Can you list 10 recombinant proteins which are used in medical practice? Find out where they are used as therapeutics.
Solution:
Recombinant proteins used in medical practice as therapeutics are as follows:
- OKT-3, a therapeutic antibody is used for reversal of acute kidney transplantation rejection.
- ReoPro is for prevention of blood clots.
- Tissue plasminogen activator (TPA) is for acute myocardial infarction.
- Asparaginase is for treatment of some types of cancer.
- DNase is for treatment of cystic fibrosis.
- Insulin is used in diabetes mellitus.
- Blood clotting factor VIII is used for treatment of haemophilia A.
- Blood clotting factor IX is for treatment of haemophilia B.
- Hepatitis B vaccine is for prevention of hepatitis B.
- Platelet derived growth factor has been approved for diabetic/skin ulcers. It also stimulates wound healing.
Question 3.
From what you have learnt, can you tell whether enzymes are bigger or DNA is bigger in molecular size? How did you know?
Solution:
Enzymes are bigger than DNA as they are proteins and proteins are macromolecules made of amino acids which are bigger than nucleotides. This can also be proved by gel electrophoresis, where denatured protein would not move but denatured DNA will move to a distance. Protein synthesis is regulated by small portions of DNA, called genes.
Question 4.
What would be the molar concentration of human DNA in a human cell? Consult your teacher.
Solution:
Molar concentration, is the ratio of the number of moles of solute in a solution divided by the volume of the solution expressed in litres.
Average weight of a DNA basepair = 650 daltons (1 dalton equals mass of a single hydrogen atom or 1.67 x 10-24 grams)
Molecular weight of a double stranded
DNA molecule = Total no. of basepairs x 650 daltons
The human genome is 3.3 x 109 bp in length.
Hence, the weight of human genome,
= 3.3 x 109 bp x 650 Da
= 3.59 x 10-12 gm.
The molar concentration of DNA can be calculated accordingly.
Question 5.
Do eukaryotic cells have restriction endonucleases? Justify your answer.
Solution:
Restriction enzymes also called ‘the molecular scissors’ are used to break DNA molecules. These enzymes are present in many bacteria where they function as a part of their defence mechanism called as restriction modification system. Molecular basis of this system was explained first by W. Arber in 1965. The restriction-modification system consists of two components;
- A restriction enzyme which identifies the introduced foreign DNA and cuts into pieces and is called restriction endonuclease,
- The second component is a modification enzyme in which methylation is done. Once a base in a DNA sequence is modified by addition of a methyl group, the restriction enzymes fail to recognise and could not cut that DNA. This is how a bacterium modifies and therefore, protects its own chromosomal DNA from cleavage by these restriction enzymes. Eukaryotic cells do not have restriction endonucleases (or restriction modification system). The DNA molecules of eukaryotes are heavily methylated by a modification enzyme, called methylase. Eukaryotes exhibit some different mechanism to counteract viral attack.
Question 6.
Besides better aeration and mixing properties, what other advantages do stirred tank bioreactors have over shake flasks?
Solution:
Shake flasks are used for growing and mixing the desired materials on a small scale in the laboratory. Bioreactors are vessels in which raw materials are biologically converted into specific products by microbes, plant and animal cells and their enzvmes. Bioreactors are used for large scale production of biomass or cell products under aseptic conditions. Here large volumes (100-1000 litres) of culture can be processed. A bioreactor provides the optimal conditions for achieving the desired product by providing optimum growth conditions (temperature, pH, substrate, salts, vitamins, oxygen). The most commonly used bioreactors are of stirring type.
A bioreactor is more advantageous than shake flasks. It has an agitator system to mix the contents properly, an oxygen delivery system to make availability of oxygen, a foam control system, a temperature control system, a pH control system and a sampling port to withdraw the small volumes of the culture periodically.
Question 7.
Collect 5 examples of palindromic DNA sequences by consulting your teacher. Better try to create a palindromic sequence by following base-pair rules.
Solution:
palindromes in DNA are base pair sequences that are the same when read forward (left to right) or backward (right to left) from a central axis of symmetry. For example, the following sequences read the same on the two strands in 5′–> 3′ direction as well as 3′ –> 5′ direction.
- 5′ – G – G – A – T – C – C – 3′
3′ – C – C – T – A – G – G – 5′ - 5′ – G – A – A – T – T – C – 3′
5′ – C – T – T – A – A – G – 5′ - 5′ – A – A – G – C – T – T – 3′
3′ – T – T – C – G – A – A – 5′ - 5′ – G – T – C – G – A – C – 3′
3′ – C – A – G – C – T – G – 5′ - 5′ – A – C – T – A – G – T – 3′
3′ – T – G – A – T – C – A – 5′
Question 8.
Can you recall meiosis and indicate at what stage a recombinant DNA is made?
Solution:
Meiosis I – Pachytene – When recombination nodule appear after synaptonemal complex formation.
Question 9.
Can you think and answer how a reporter enzyme can be used to monitor transformation of host cells by foreign DNA in addition to a selectable marker?
Solution:
Transformation is a process through which a piece of DNA is introduced into a host bacterial cell. Normally, the genes encoding resistance to antibiotics such as ampicillin, tetracycline, etc., are considered useful selectable markers to differentiate between transformed and non-transformed bacterial cell. In addition to these selectable markers, an alternative selectable marker has been developed to differentiate transformed and non-transformed bacterial cell on the basis of their ability to produce colour in the presence of a chromogenic substance. A recombinant DNA is inserted in the coding sequence of an enzyme (5-galactosidase (reporter enzyme). If the plasmid in the bacterium does not have an insert, the presence of a chromogenic substance gives blue coloured colonies, presence of insert results into insertional inactivation of (3-galactosidase and, therefore, the colonies do not produce any colour, these colonies are marked as transformed colonies.
Question 10.
Describe briefly the following :
(a) Origin of replication
(b) Bioreactors
(c) Downstream processing
Solution:
(a) Origin of replication (on): One of the major components of a plasmid is a sequence of bases where replication starts. It is called origin of replication (on). This is a specific portion of plasmid genome that serves as start signal for self-replication (to make another copy of itself). Any piece of DNA when linked to this sequence can be made to replicate within the host cells. This property is used to make a number of copies of linked DNA (or DNA insert).
(b) Bioreactors : These are vessels in which raw materials are biologically converted into specific products by microbes, plant and
animal cells and their enzymes. They are allowed to synthesise the desired proteins which are finally extracted and purified from cultures. Small volume cultures are usually employed in laboratories for research and production of less quantities of products. However, large scale production of the products is carried out in ‘bioreactors’. The most commonly used bioreactors is stirring type bioreactor (fermenter) that has a provision for batch culture or continuous culture.
(c) Downstream processing : After the formation of the product in the bioreactors, it undergoes through some processes before a finished product is ready for marketing. These processes include separation and purification of products which are collectively called the downstream processing. The product is then subjected to quality control testing and kept in suitable preservatives. The downstream process and quality control test are different for different products
Question 11.
Explain briefly
(a) PCR
(b) Restriction enzymes and DNA
(c) Chitinase
Solution:
(a) Polymerase chain reaction (PCR) is a technique of synthesizing multiple copies of the desired gene (DNA) in vitro. This technique was developed by Kary Mullis in 1985. It is based on the principle that a DNA molecule, when subjected to high temperature, splits into two strands due to denaturation. These single stranded molecules are then converted to double stranded molecules by synthesising new strands in presence of enzyme DNA polymerase. Thus, multiple copies of the original DNA sequence can be generated by repeating the process several times. The basic requirements of PCR are, DNA template, two nucleotide primers usually 20 nucleotides long and enzyme DNA polymerase which is stable at high temperature (usually Taq polymerase):
Working mechanism of PCR is as follows:
- First of all, the target DNA (DNA segment to be amplified) is heated to high temperature (94 to 96° C). Heating results in the separation of two strands of DNA. Each of the two strands of the target DNA now acts as template for synthesis of new DNA strand. This step is called denaturation.
- Denaturation is followed by annealing. During this step, two oligonucleotide primers hybridise to each of single stranded template DNA in presence of excess of synthetic oligonucleotides. Annealing is carried out at lower temperature (40° – 60°C).
- Third and final step is extension. During this step, the enzyme DNA polymerase synthesises the DNA segment between the primers. Usually Taq DNA polymerase, isolated from a thermophilic bacterium Thermus acquaticus, is used in most of the cases. The two primers extend towards each other in order to copy the DNA segment lying between the two primers. This step requires presence of deoxynucleoside triphosphates (dNTPs) and Mg2+ and occurs at 72°C.
The above mentioned three steps complete the first cycle of PCR. The second cycle begins with denaturation of extension product of first cycle and after completing the extension step, two cycles are completed. If these cycles are repeated many times, the DNA segment can be amplified to approximately billion times, i.e., one billion copies of desired DNA segment are made.
(b) Restriction enzymes are used to break DNA molecules. They belong to a larger class of enzymes called nucleases. Restriction enzymes are of three types – exonucleases, endonucleases and restriction endonucleases,
- Exonucleases : They remove nucleotides from the terminal ends (either 5′ or 3′) of DNA in one strand of duplex.
- Endonucleases: They make cuts at specific position within the DNA. These enzymes do not cleave the ends and involve only one strand of the DNA duplex.
- Restriction endonucleases : These were found by Arber in 1963 in bacteria. They act as “molecular scissors” or chemical scalpels. They recognise the base sequence at palindrome sites in DNA duplex and cut its strands. Three main types of restriction endonucleases are type I, type II and type III. Out of the three types, only type II restriction enzymes are used in recombinant DNA technology because they can be used in vitro to recognise and cut within specific DNA sequence typically consisting of 4 to 8 nucleotides.
(c) Chitinase is a lysing enzyme that dissolves the fungal cell wall. It results in the release of DNA along with several other macro molecules.
Studyit.in is the best & trusted site that offers all study materials and exam preparation resources for CBSE Class 1 to Class 12 students. So, you can find class 12 NCERT Solutions for Biology from this website. If you liked this solution, please do so and share it among your classmates so that more students can benefit from it. Visit for CBSE Biology class 12 Solutions Chapter wise PDF notes of all classes and all subjects are available here
Some Useful Links For CBSE Class 12



0 Comments
Please Comment